Consistent quality, always delivered within 3 days.
Actual scientists responding to you within 24 hours.
All the data you need to evaluate your results.
Get ready for a new type of DNA Sequencing service.
For scientists, by scientists.
Whole Plasmid Sequencing
Insert orientation
See the orientation of your plasmid to make sure your ORFs are expressed as intended.
gDNA contamination score
We regularly see customer samples where only 10% of the sample is actually plasmid DNA.
No primers
Say goodbye to primer design and annealing issues. We just need your DNA, nothing else.
Full plasmid validation
Is the ori correct? Is your marker not working as expected? Check your entire plasmid to find out.
Complex regions
Validate repetitive and complex regions larger than the max read length of Sanger or short-read NGS.
Cheaper than Sanger
115 DKK and one single reaction for plasmids <25kb makes us cheaper than Sanger for large ROIs.
How does it work?
Measure your samples
Register your order and measure concentration and purity of your DNA samples, as described in our submission guide . We require at least 20 ul of DNA at 5.6 ng/ul with high purity.

Submit your samples
Transfer your samples to either PCR tubes or 96-well plates and prepare them for secure shipment . We have no minimum sample number and you can submit samples via mail or Dropboxes at your institution. Not sure if your institute already has a dropbox? Just reach out to us here.

We do the sequencing
Your samples are processed by our experienced laboratory staff which operate our automated robotics workflow to prepare samples for nanopore sequencing. The generated sequencing reads are then run through our proprietary bioinformatics pipeline to generate your final plasmid assemblies.
You enjoy the results
Your results are ready for download from our website within 3 days. You will receive a fully annotated plasmid assembly together with a detailed quality report. Raw reads are available upon request.
Have confidence in your results
-
Annotated quality PDF
-
Annotated Genbank
-
Read length plot
-
Edible plasmids
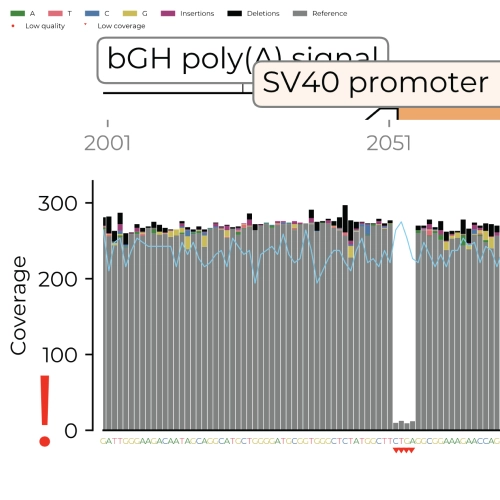
Low quality positions highlighted
The Quality PDF shows 3 quality metrics for each nucleotide position:
- Read coverage (Grey bars)
- FASTQ PHRED quality score (Cyan line)
- Nucleotide variants (Coloured bars)
Below-threshold positions are marked with red symbols.

Annotations for below threshold positions
Annotations for below-threshold QC positions are shown in your genbank file. Search for:
- "LOW-QUALITY"
- "POSSIBLE-VARIANT"
- "LOW-COVERAGE"
Simply align the gbk to your reference and see which nucleotide positions need further investigation.
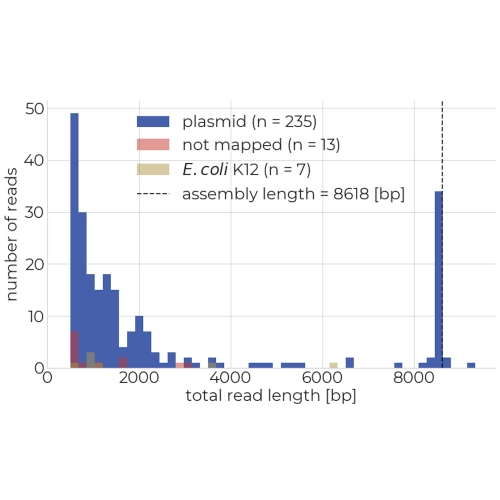
Origin and length of sequencing reads
A read length histogram shows the length of your sequencing reads and whether they mapped to your plasmid assembly, the E. coli K12 genome, or neither. You can also determine the extend of fragmentation based on the number of short reads.
Edible plasmids for our most loyal customers
The trust of our customers means everything to us. So when we see that scientists stick with us for years, we try to show it every way we can, including edible plasmids (Donuts).
Finally, PhD-level customer support!
Before, during, and after sequencing: Our PhD-level scientists are here to discuss new projects, answer any questions, or help with improving sequencing success.
All within 24 hours of reaching out to us.

What our customers say
“Unveil Bio's plasmid sequencing service was thorough, timely, and of high quality. The detailed submission guide provided on the website makes the submission process extremely easy. Retrieving sequencing data is effortless via the user-friendly website. Awesome work!”
“I wanted to say thank you for our collaboration. I have really enjoyed working with you and implementing your service in our company. You and your team has been super service minded and I have enjoyed that your input has always been given with a problem-solving attitude."
"Unveil Bio's Whole Plasmid Sequencing service is a game changer for lab geeks! It was affordable and of better quality than Sanger sequencing. Sequencing the whole plasmid was much more convenient than just the gene especially when it is super long - saved me a lot of time and hassle."
Our services
Whole Plasmid Sequencing
115 DKK/sample
Submitted at 5.6 ng/ul in > 20 ul. For monoclonal plasmids of 2.5 - 25 kb
Sequenced with Nanopore Technology using a PCR-free method.
-
Pick up from sample dropbox
-
Personalised results email
-
Extensive results files
-
Turnaround within 3 days
-
Up to 20% volume discounts
Whole Amplicon Sequencing
115 DKK/sample
Submitted as 5 ul of un-purified un-diluted PCR reaction. For amplicons of 0.7 - 10 kb.
Sequenced with Nanopore Technology using a PCR-free method.
-
Pick up from sample dropbox
-
Personalised results email
-
Extensive results files
-
Turnaround within 3 days
-
Up to 20% volume discounts
Custom Projects
Inquire
From raw reads generation to custom bioinformatic analysis, we are always happy to discuss custom projects.
Get in touch and tell us about your sequencing needs.
More questions?
How are samples sequenced?
We use Nanopore Technology (ONT) long-read sequencing technology to sequence our customer samples. We first construct a sequencing library using ONT Kit v14 library preparation chemistry. During this step, circular plasmid DNA is linearized in a sequence-independent manner. The library is then sequenced, without primers, on R10.4.1 flow cells with raw data accuracy >99%. As a last step the raw reads are run through our bioinformatics pipeline to create a high accuracy assembly that is fully annotated. All samples are sequenced without any amplification steps or primers. Hence, please avoid sending primers along or inside your samples.
How should I ship my samples to you?
You can either send your samples to us directly or use one of our dropboxes at your institution. Sequencing is available throughout the week - Tuesday to Friday. Please closely follow our Submission Guide for details on sample preparation and shipping.
How accurate is nanopore sequencing?
Oxford Nanopore Technology has come a long way and now enables high accuracy comparable to Sanger sequencing:
- > 99% raw read accuracy
- > 99.999% / Q50 consensus accuracy
What data files will I receive?
- .fasta file: A polished consensus sequence of the plasmid in fasta format.
- .gbk GenBank file: An annotated pLannotate plasmid map in GenBank format. Positions below our QC thresholds are marked as annotation and indicate low confidence in the accuracy of the indicated base; LOW-QUALITY, LOW_COVERAGE, POSSIBLE-VARIANT.
- quality.pdf file: A quality report in pdf format that shows the number of times a base has been sequenced in grey bars (Coverage) and a quality score for each base position as a cyan coloured line (FASTQ PHRED Quality). The latter is an internal quality metric where high values indicate a high confidence in the accuracy of the identified base. In addition, the quality report shows nucleotide variants that were found at each sequence position and their abundance is indicated by the height of the coloured bar. E.g.: If sequence position 256 has a grey bar for base T and 30% of the bar is coloured green then 30% of the sequence reads had an A at this position and 70% had a T. Bases that represent >60% of all base variants found are colored in grey.
- quality.tsv: The same information as in the quality.pdf file but in tsv format which can be opened in Excel or similar
- read lenght plot: A histogram showing the number of sequencing reads and their length. Bar colors indicate whether the reads mapped to the assembly, E. coli chromosome, or neither. The dotted vertical line indicates the assembly size. Ideally, the majority of sequencing reads should have a legnth equal to the assemly length which indicates that the entire plasmid was sequences in single continuous reads.
- .html pLannotate map: An annotated plasmid map, generated using the pLannotate tool from the Barrick Lab.